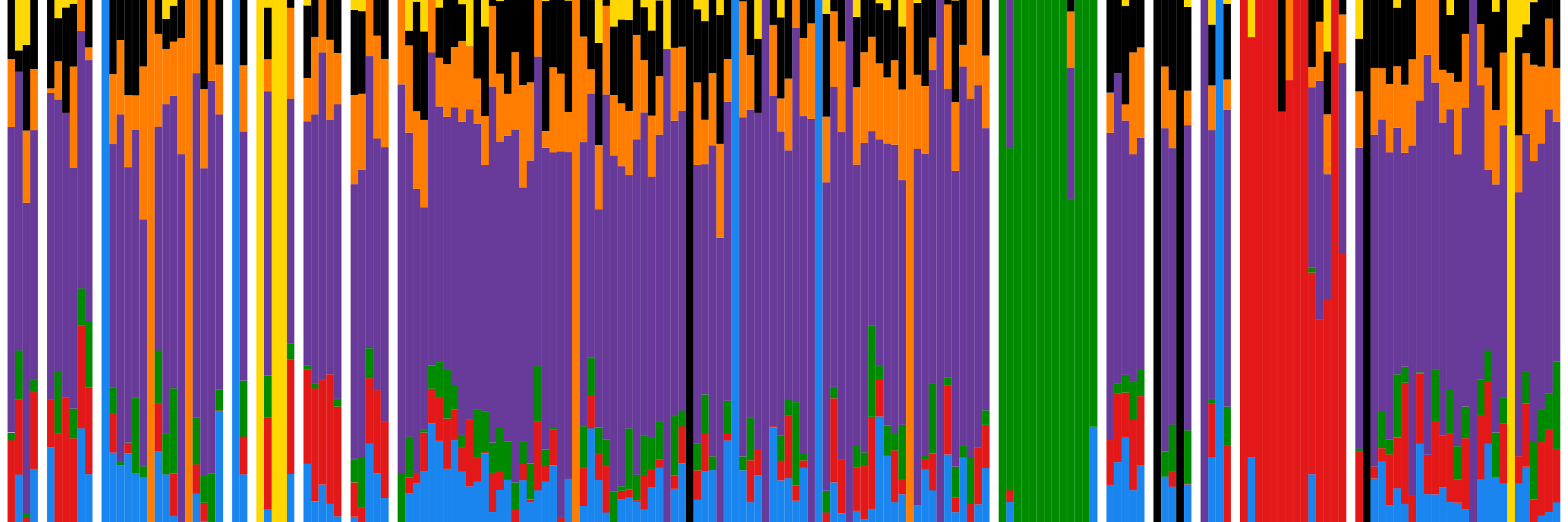
The general topics that the Genomics of Individuality lab is interested in revolve around the genomics of individuality: What is there in our genomes that makes us the way we are? What does it tell us about our ancestry? How does it affect our susceptibility to diseases? How can this be applied in practical settings (i.e., in forensic genetics)? In particular, its research focuses on populations on both sides of the western Mediterranean, looking at genetic structure, and contacts, and, in particular, populations that are geographically or socially isolated.
Lab website: Calafell Lab